GROMACS + AMD EPYC: High-Performance Molecular Dynamics
GROMACS is a powerful open-source molecular dynamics package primarily designed for simulations of proteins, lipids, nucleic acids, as well as non-biological systems such as polymers. GROMACS supports all the usual algorithms expected from a modern molecular dynamics implementation. GROMACS can be run in parallel in a multi-node environment using the standard MPI communication protocol, and since GROMACS 4.6, has implemented CUDA-based GPU acceleration on NVIDIA GPUs. However, this blog will focus on the CPU performance of GROMACS on AMD EPYC 7002 Processors
GROMACS is known for being extremely user-friendly, with topologies and parameter files written in clear text format. With GROMACS there is a lot of consistency checking, and clear error messages are issued when errors arise.
AMD’s new 7002 Series server processors are all the rage at the moment, as the new 7002's feature up to 64 ‘Zen 2’ cores per SoC, and deliver vast improvements on per-core server workloads over several benchmarks. In a recent whitepaper by AMD, this proves the case as they ran workloads on GROMACS vs Gen1 EPYC processors.
GROMACS Performance: 1st Gen vs 2nd Gen EPYC Processors
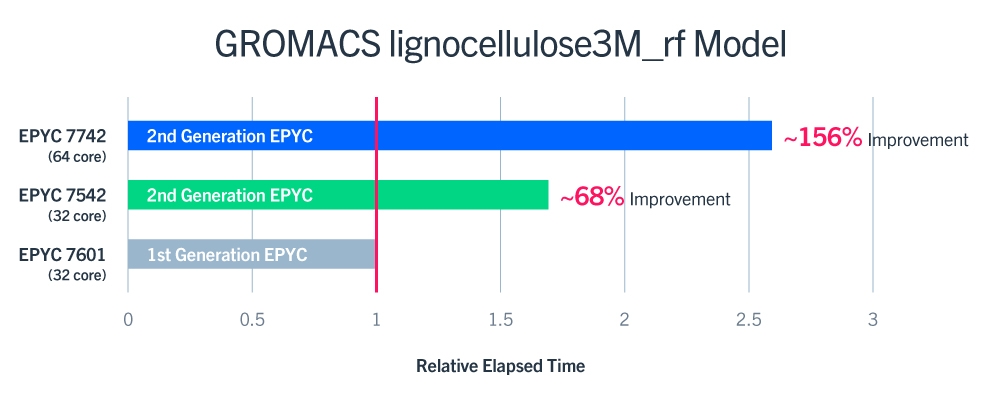
In the aforementioned whitepaper, it details the single-node performance of two-socket systems across each of the specific AMD EPYC processors (32 core 1st Gen 7601, 32 core 2nd Gen 7542, and 64 core 2nd gen 7742). The results show a very strong improvement of nearly 68% between the EPYC 7601 and the EPYC 7542. Furthermore, the results show that GROMACS can take advantage of the higher core count of the 2nd Gen EPYC 7742, with an impressive ~156% average performance boost over the 1st generation EPYC 7601! According to the paper, multi-threading was disabled in the cluster and the job was run in a pure MPI configuration during these tests.
GROMACS: Multi-node Scaling with EPYC Processors
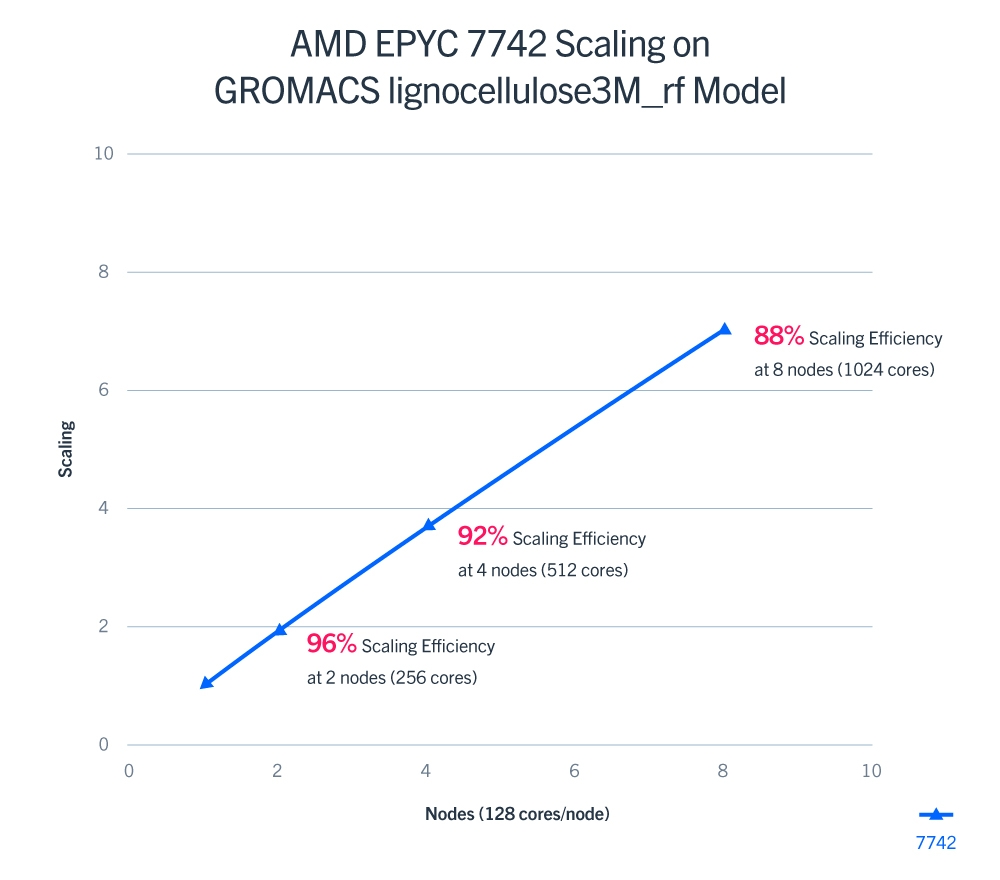
AMD 2nd Gen EPYC processors and showed incredible generational performance gains over the AMD 1st Gen EPYC 7601. The 64-core EPYC 7742 showed an ~156% performance increase and the 32-core EPYC 7542 showed an ~68% performance increase over the prior generation. Scaling testing was then run on the lignocellulose3M_rf model from PRACE to show the scaling of GROMACS on the EPYC 7742. Results show very good scaling of ~96%, ~92%, and ~88% at 2, 4, and 8 nodes, respectively.
Take GROMACS Simulations to the next level with GPUs
Combine the latest EPYC Processors with CUDA enabled NVIDIA GPUs to get the most out of your MD Simulations. By leveraging the CUDA parallel processing architecture of NVIDIA GPUs, GROMACS CUDA GPU acceleration is now a core part of GROMACS that works in combination with GROMACS' domain decomposition and load balancing code, delivering performance up to massive performance gains when compared to CPU-only processing. Exxact has teamed up with the GROMACS development team to design a series of certified GROMACS Servers and Workstations that provide optimum price and performance for a wide range of research activities.
4U GPU HPC Node for GROMACS
[supsystic-tables id=55]
Other Molecular Dynamics Related Blogs
- GPU Accelerated Cryo-EM using RELION: From Workstation to Supercomputer
- A Summary of DeepMind’s Protein Folding Upset at CASP13
- Deep Learning Meets Molecular Dynamics: “Predicting Correctness of Protein Complex Binding Operations”- An Interview With Stanford Students

Molecular Dynamics Simulation with GROMACS on AMD EPYC 7002 Powered Servers
GROMACS + AMD EPYC: High-Performance Molecular Dynamics
GROMACS is a powerful open-source molecular dynamics package primarily designed for simulations of proteins, lipids, nucleic acids, as well as non-biological systems such as polymers. GROMACS supports all the usual algorithms expected from a modern molecular dynamics implementation. GROMACS can be run in parallel in a multi-node environment using the standard MPI communication protocol, and since GROMACS 4.6, has implemented CUDA-based GPU acceleration on NVIDIA GPUs. However, this blog will focus on the CPU performance of GROMACS on AMD EPYC 7002 Processors
GROMACS is known for being extremely user-friendly, with topologies and parameter files written in clear text format. With GROMACS there is a lot of consistency checking, and clear error messages are issued when errors arise.
AMD’s new 7002 Series server processors are all the rage at the moment, as the new 7002's feature up to 64 ‘Zen 2’ cores per SoC, and deliver vast improvements on per-core server workloads over several benchmarks. In a recent whitepaper by AMD, this proves the case as they ran workloads on GROMACS vs Gen1 EPYC processors.
GROMACS Performance: 1st Gen vs 2nd Gen EPYC Processors
In the aforementioned whitepaper, it details the single-node performance of two-socket systems across each of the specific AMD EPYC processors (32 core 1st Gen 7601, 32 core 2nd Gen 7542, and 64 core 2nd gen 7742). The results show a very strong improvement of nearly 68% between the EPYC 7601 and the EPYC 7542. Furthermore, the results show that GROMACS can take advantage of the higher core count of the 2nd Gen EPYC 7742, with an impressive ~156% average performance boost over the 1st generation EPYC 7601! According to the paper, multi-threading was disabled in the cluster and the job was run in a pure MPI configuration during these tests.

GROMACS: Multi-node Scaling with EPYC Processors
AMD 2nd Gen EPYC processors and showed incredible generational performance gains over the AMD 1st Gen EPYC 7601. The 64-core EPYC 7742 showed an ~156% performance increase and the 32-core EPYC 7542 showed an ~68% performance increase over the prior generation. Scaling testing was then run on the lignocellulose3M_rf model from PRACE to show the scaling of GROMACS on the EPYC 7742. Results show very good scaling of ~96%, ~92%, and ~88% at 2, 4, and 8 nodes, respectively.

Take GROMACS Simulations to the next level with GPUs
Combine the latest EPYC Processors with CUDA enabled NVIDIA GPUs to get the most out of your MD Simulations. By leveraging the CUDA parallel processing architecture of NVIDIA GPUs, GROMACS CUDA GPU acceleration is now a core part of GROMACS that works in combination with GROMACS' domain decomposition and load balancing code, delivering performance up to massive performance gains when compared to CPU-only processing. Exxact has teamed up with the GROMACS development team to design a series of certified GROMACS Servers and Workstations that provide optimum price and performance for a wide range of research activities.
4U GPU HPC Node for GROMACS
[supsystic-tables id=55]
Other Molecular Dynamics Related Blogs
- GPU Accelerated Cryo-EM using RELION: From Workstation to Supercomputer
- A Summary of DeepMind’s Protein Folding Upset at CASP13
- Deep Learning Meets Molecular Dynamics: “Predicting Correctness of Protein Complex Binding Operations”- An Interview With Stanford Students





.jpg?format=webp)