RTX 3090 HPC Benchmarks for RELION
Overview
As a leading supplier of scientific workstations and servers, Exxact has conducted benchmarks for RELION Cryo-EM on the new NVIDIA GeForce RTX 3090 GPUs. The results are quite good, especially when paired with an AMD EPYC 7552 Processor, we were able to run the plasmodium ribosome benchmark in under an hour.
Interested in getting faster Cryo-EM results?
Learn more about RELION GPU Accelerated Systems for Cryo-EM
RELION GPU Support Summary
With advancements in automation, compute power, and visual technology, the scope and complexity of datasets used in cryo-EM have grown substantially. GPU support and acceleration are essential for the flexibility of resource management, prevention of memory limitations, and to address the most computationally intensive processes of cryo-EM such as image classification, and high-resolution refinement.
System Specs
| Base System Configuration | |
| Nodes | 1 |
| Processor | AMD EPYC 7552 |
| Processor Count | 2 |
| Total Logical Cores | 48 |
| Memory Type | DDR4 |
| Memory Size | 512 GB |
| Storage | SSD |
| OS | CentOS 7 |
| CUDA Version | 10.2 |
| Relion Version | 3 |
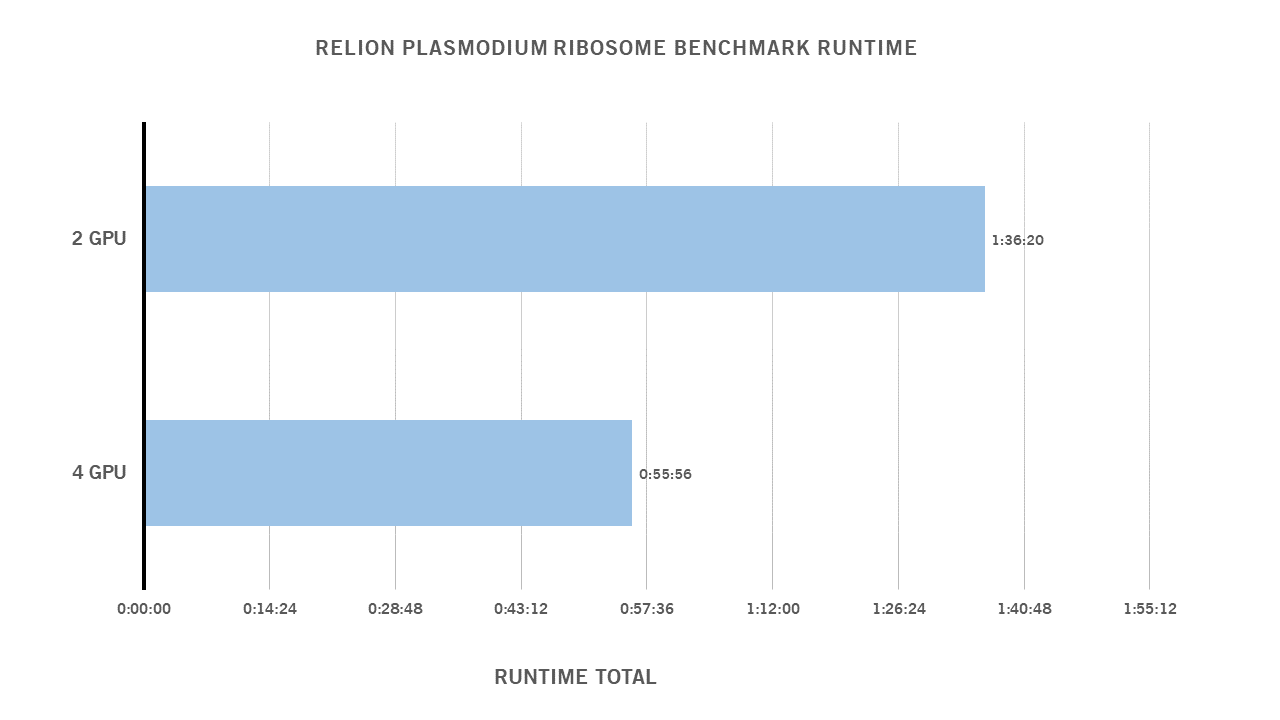
RTX 3090 Relion Benchmarks 2GPU & 4 GPU Configuration
Plasmodium ribosome data set ftp://ftp.mrc-lmb.cam.ac.uk/pub/scheres/relion_benchmark.tar.gz
Benchmark Parameters 4 GPU
<ul>mpirun -n 5 /usr/local/relion-3/bin/relion_refine_mpi --j 6 --gpu --pool 100 --dont_combine_weights_via_disc --keep_scratch --reuse_scratch --i Particles/shiny_2sets.star --ref emd_2660.map:mrc --firstiter_cc --ini_high 60 --ctf --ctf_corrected_ref --iter 25 --tau2_fudge 4 --particle_diameter 360 --K 6 --flatten_solvent --zero_mask --oversampling 1 --healpix_order 2 --offset_range 5 --offset_step 2 --sym C1 --norm --scale --random_seed 0 --o class3d</ul>Benchmark Parameters 2 GPU
<ul>mpirun -n 3 /usr/local/relion-3/bin/relion_refine_mpi --j 6 --gpu --pool 100 --dont_combine_weights_via_disc --keep_scratch --reuse_scratch --i Particles/shiny_2sets.star --ref emd_2660.map:mrc --firstiter_cc --ini_high 60 --ctf --ctf_corrected_ref --iter 25 --tau2_fudge 4 --particle_diameter 360 --K 6 --flatten_solvent --zero_mask --oversampling 1 --healpix_order 2 --offset_range 5 --offset_step 2 --sym C1 --norm --scale --random_seed 0 --o class3d</ul>Notes on System Memory
Although a minimum of 64 GB of RAM is recommended to run RELION with small image sizes (eg. 200×200) on either the original or accelerated versions of RELION, 360×360 problems run best on systems with more than 128GB of RAM. Systems with 256GB or more RAM are recommended for the CPU-accelerated kernels on larger image sizes. Insufficient memory causes individual MPI ranks to be killed, leading to zombie RELION jobs.
MPI Settings
Where some users may want to run more than one MPI rank per GPU, sufficient GPU memory is needed. Each MPI-slave that shares a GPU increases the use of memory. In this case, however, it’s recommended running a single MPI-slave per GPU for good performance and stable execution.
Notes on Scaling
The GPUs tested were Turing/Volta-based and performed similarly. As a result, it is more beneficial to scale out than scale up. Another thing to note is the diminishing returns in scaling once you pass 4 GPUs.
Have any questions about RELION or other applications for molecular dynamics?
Contact Exxact Today

NVIDIA RTX 3090 RELION Cryo-EM Benchmarks and Analysis
RTX 3090 HPC Benchmarks for RELION
Overview
As a leading supplier of scientific workstations and servers, Exxact has conducted benchmarks for RELION Cryo-EM on the new NVIDIA GeForce RTX 3090 GPUs. The results are quite good, especially when paired with an AMD EPYC 7552 Processor, we were able to run the plasmodium ribosome benchmark in under an hour.
Interested in getting faster Cryo-EM results?
Learn more about RELION GPU Accelerated Systems for Cryo-EM
RELION GPU Support Summary
With advancements in automation, compute power, and visual technology, the scope and complexity of datasets used in cryo-EM have grown substantially. GPU support and acceleration are essential for the flexibility of resource management, prevention of memory limitations, and to address the most computationally intensive processes of cryo-EM such as image classification, and high-resolution refinement.
System Specs
| Base System Configuration | |
| Nodes | 1 |
| Processor | AMD EPYC 7552 |
| Processor Count | 2 |
| Total Logical Cores | 48 |
| Memory Type | DDR4 |
| Memory Size | 512 GB |
| Storage | SSD |
| OS | CentOS 7 |
| CUDA Version | 10.2 |
| Relion Version | 3 |
RTX 3090 Relion Benchmarks 2GPU & 4 GPU Configuration
Plasmodium ribosome data set ftp://ftp.mrc-lmb.cam.ac.uk/pub/scheres/relion_benchmark.tar.gz
Benchmark Parameters 4 GPU
<ul>mpirun -n 5 /usr/local/relion-3/bin/relion_refine_mpi --j 6 --gpu --pool 100 --dont_combine_weights_via_disc --keep_scratch --reuse_scratch --i Particles/shiny_2sets.star --ref emd_2660.map:mrc --firstiter_cc --ini_high 60 --ctf --ctf_corrected_ref --iter 25 --tau2_fudge 4 --particle_diameter 360 --K 6 --flatten_solvent --zero_mask --oversampling 1 --healpix_order 2 --offset_range 5 --offset_step 2 --sym C1 --norm --scale --random_seed 0 --o class3d</ul>Benchmark Parameters 2 GPU
<ul>mpirun -n 3 /usr/local/relion-3/bin/relion_refine_mpi --j 6 --gpu --pool 100 --dont_combine_weights_via_disc --keep_scratch --reuse_scratch --i Particles/shiny_2sets.star --ref emd_2660.map:mrc --firstiter_cc --ini_high 60 --ctf --ctf_corrected_ref --iter 25 --tau2_fudge 4 --particle_diameter 360 --K 6 --flatten_solvent --zero_mask --oversampling 1 --healpix_order 2 --offset_range 5 --offset_step 2 --sym C1 --norm --scale --random_seed 0 --o class3d</ul>Notes on System Memory
Although a minimum of 64 GB of RAM is recommended to run RELION with small image sizes (eg. 200×200) on either the original or accelerated versions of RELION, 360×360 problems run best on systems with more than 128GB of RAM. Systems with 256GB or more RAM are recommended for the CPU-accelerated kernels on larger image sizes. Insufficient memory causes individual MPI ranks to be killed, leading to zombie RELION jobs.
MPI Settings
Where some users may want to run more than one MPI rank per GPU, sufficient GPU memory is needed. Each MPI-slave that shares a GPU increases the use of memory. In this case, however, it’s recommended running a single MPI-slave per GPU for good performance and stable execution.
Notes on Scaling
The GPUs tested were Turing/Volta-based and performed similarly. As a result, it is more beneficial to scale out than scale up. Another thing to note is the diminishing returns in scaling once you pass 4 GPUs.
Have any questions about RELION or other applications for molecular dynamics?
Contact Exxact Today