Performance Benchmarks for RELION
Overview
As a value-added supplier of scientific workstations and servers, Exxact regularly provides reference benchmarks in various GPU configurations to guide Cryogenic electron microscopy (cryo-EM) scientists looking to procure systems optimized for their research.
Software Summary
RELION (REgularised LIkelihood OptimisatioN), or Relion, has revolutionized the cryo–EM field since 2012. Developed by Scheres Lab at the MRC Laboratory of Molecular Biology, this stand-alone computer program uses a Bayesian approach to refine macromolecular structures by single-particle analysis of electron cryo-microscopy data.
The development of RELION is supported through long-term funding by the UK Medical Research Council, and is distributed under a GPLv2 license. This means that anyone (including commercial users) can download, use and modify RELION without having to pay anything. They just request that if RELION is useful in your work, that you cite their papers
Interested in getting faster results?
Learn more about RELION GPU Accelerated Systems for Cryo-EM starting at $9,599
RELION GPU Support Summary
With advancements in automation, compute power, and visual technology, the scope and complexity of datasets used in cryo-EM have grown substantially. GPU support and acceleration are essential for the flexibility of resource management, prevention of memory limitations, and to address the most computationally intensive processes of cryo-EM such as image classification, and high-resolution refinement.
System Specs
| Base System Configuration | |
| Make | Exxact |
| Model | TS4-1709194-REL |
| Nodes | 1 |
| Processor | Intel Xeon Silver 4116 |
| Processor Count | 2 |
| Total Logical Cores | 48 |
| Memory Type | DDR4 |
| Memory Size | 128 GB |
| Storage | SSD |
| OS | CentOS 7 |
| CUDA Version | 10.2 |
| Relion Version | 3 |
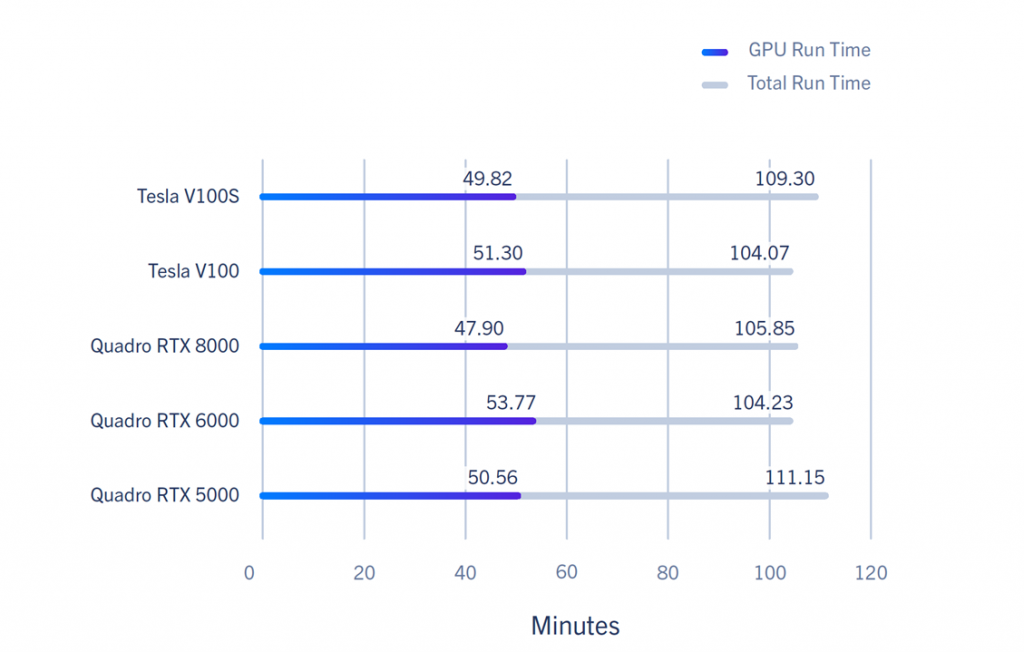
RELION Benchmarks – GPU Run Time Performance
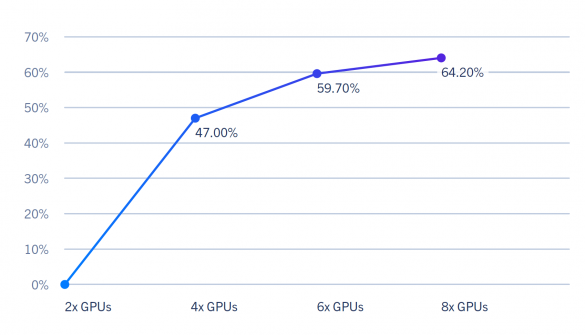
RELION Benchmarks – GPU Speedup Across Multiple GPUs (NVIDIA Quadro RTX 8000)
Notes on System Memory
Although a minimum of 64 GB of RAM is recommended to run RELION with small image sizes (eg. 200×200) on either the original or accelerated versions of RELION, 360×360 problems run best on systems with more than 128GB of RAM. Systems with 256GB or more RAM are recommended for the CPU-accelerated kernels on larger image sizes. Insufficient memory causes individual MPI ranks to be killed, leading to zombie RELION jobs.
MPI Settings
Where some users may want to run more than one MPI rank per GPU, sufficient GPU memory is needed. Each MPI-slave that shares a GPU increases the use of memory. In this case, however, it’s recommended running a single MPI-slave per GPU for good performance and stable execution.
Notes on Scaling
The GPUs tested were Turing/Volta-based and performed similarly. As a result, it is more beneficial to scale out than scale up. Another thing to note is the diminishing returns in scaling once you pass 4 GPUs.
Have any questions about RELION or other applications for molecular dynamics?
Contact Exxact Today

RELION Cryo-EM Benchmarks and Analysis
Performance Benchmarks for RELION
Overview
As a value-added supplier of scientific workstations and servers, Exxact regularly provides reference benchmarks in various GPU configurations to guide Cryogenic electron microscopy (cryo-EM) scientists looking to procure systems optimized for their research.
Software Summary
RELION (REgularised LIkelihood OptimisatioN), or Relion, has revolutionized the cryo–EM field since 2012. Developed by Scheres Lab at the MRC Laboratory of Molecular Biology, this stand-alone computer program uses a Bayesian approach to refine macromolecular structures by single-particle analysis of electron cryo-microscopy data.
The development of RELION is supported through long-term funding by the UK Medical Research Council, and is distributed under a GPLv2 license. This means that anyone (including commercial users) can download, use and modify RELION without having to pay anything. They just request that if RELION is useful in your work, that you cite their papers
Interested in getting faster results?
Learn more about RELION GPU Accelerated Systems for Cryo-EM starting at $9,599
RELION GPU Support Summary
With advancements in automation, compute power, and visual technology, the scope and complexity of datasets used in cryo-EM have grown substantially. GPU support and acceleration are essential for the flexibility of resource management, prevention of memory limitations, and to address the most computationally intensive processes of cryo-EM such as image classification, and high-resolution refinement.
System Specs
| Base System Configuration | |
| Make | Exxact |
| Model | TS4-1709194-REL |
| Nodes | 1 |
| Processor | Intel Xeon Silver 4116 |
| Processor Count | 2 |
| Total Logical Cores | 48 |
| Memory Type | DDR4 |
| Memory Size | 128 GB |
| Storage | SSD |
| OS | CentOS 7 |
| CUDA Version | 10.2 |
| Relion Version | 3 |
RELION Benchmarks – GPU Run Time Performance
RELION Benchmarks – GPU Speedup Across Multiple GPUs (NVIDIA Quadro RTX 8000)
Notes on System Memory
Although a minimum of 64 GB of RAM is recommended to run RELION with small image sizes (eg. 200×200) on either the original or accelerated versions of RELION, 360×360 problems run best on systems with more than 128GB of RAM. Systems with 256GB or more RAM are recommended for the CPU-accelerated kernels on larger image sizes. Insufficient memory causes individual MPI ranks to be killed, leading to zombie RELION jobs.
MPI Settings
Where some users may want to run more than one MPI rank per GPU, sufficient GPU memory is needed. Each MPI-slave that shares a GPU increases the use of memory. In this case, however, it’s recommended running a single MPI-slave per GPU for good performance and stable execution.
Notes on Scaling
The GPUs tested were Turing/Volta-based and performed similarly. As a result, it is more beneficial to scale out than scale up. Another thing to note is the diminishing returns in scaling once you pass 4 GPUs.
Have any questions about RELION or other applications for molecular dynamics?
Contact Exxact Today




.jpg?format=webp)