
CryoSPARC Overview
CryoSPARC (Cryo-EM Single Particle Ab-initio Reconstruction and Classification) is a software package for processing cryo-electron microscopy (cryo-EM) single particle data, used in research and drug discovery.
As a complete solution for cryo-EM processing, cryoSPARC allows:
- Ultra-fast end-to-end processing of raw cryo-EM data and reconstruction of electron density maps, ready for ingestion into model building software
- Optimized algorithms and GPU acceleration at all stages, from pre-processing through particle picking, 2D particle classification, 3D ab-initio structure determination, high resolution refinement, and heterogeneity analysis
- Specialized and unique tools for therapeutically relevant targets, membrane proteins, continuously flexible structure
- Interactive, visual and iterative experimentation for even the most complex workflows
Interested in getting faster results?
Learn more about cryoSPARC Optimized GPU Workstations
CryoSPARC v3.3 Release
The newest update is packed with new features, performance optimizations and various stability enhancements.
Download the latest version here.
We highly recommend making a backup of your cryoSPARC database before updating.
Update instructions are available here.
What's New
- New 3D Classification BETA job can be used to quickly classify particles into many 3D classes, without re-alignment of particle poses and shifts. Sort out heterogeneity from million+ particle datasets with 100+ classes. Read a tutorial here.
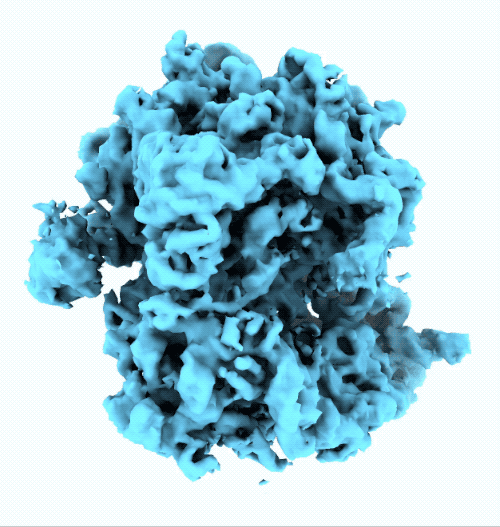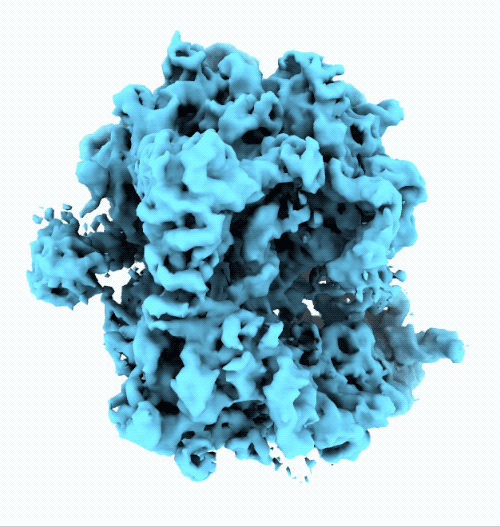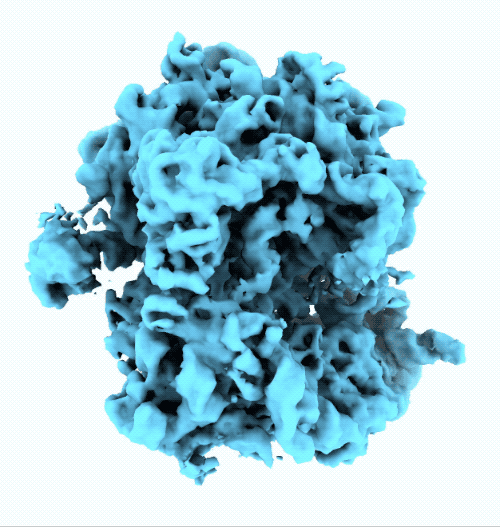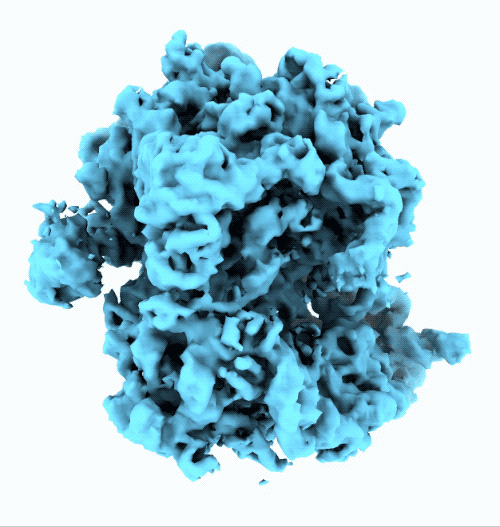
- New ruler and scale bar allow for interactively determining the size of a particle in cryoSPARC Live.
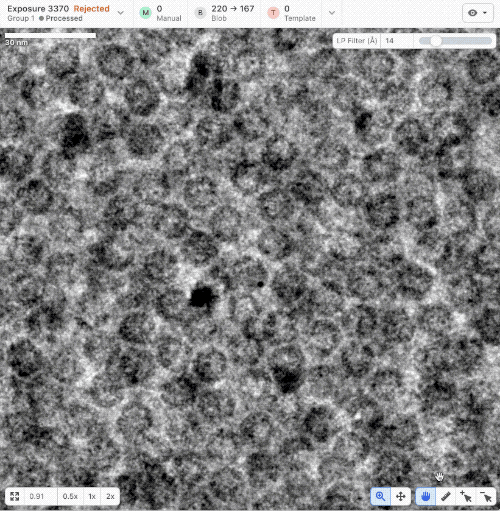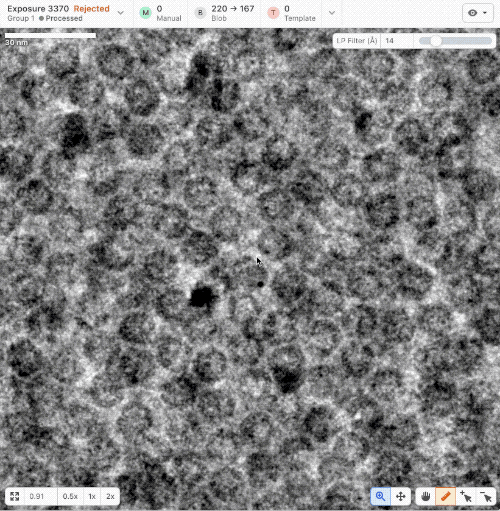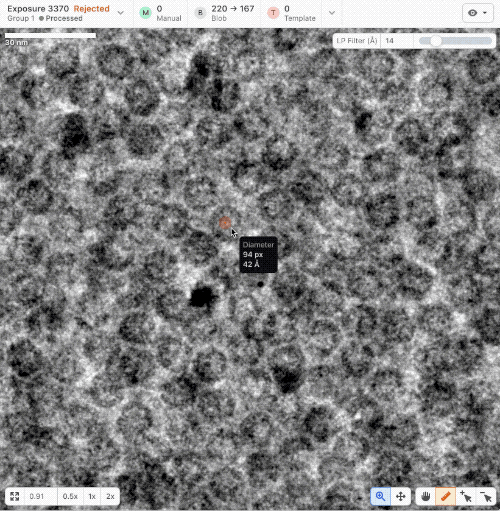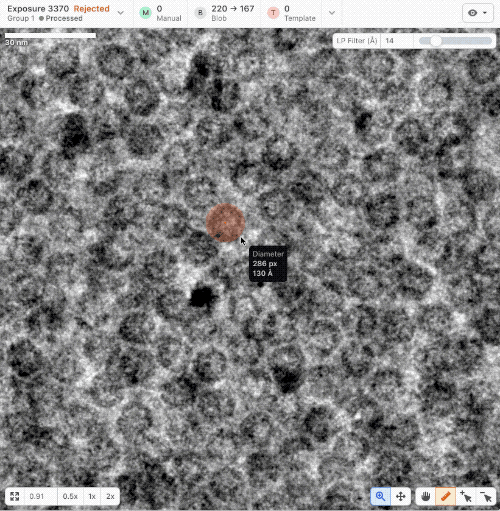
- New Blob Picker Tuner determines optimal blob picker parameters from a minimal set of user-selected manual picks. Tunes blob size, shape, and picker score thresholds automatically. Read a tutorial here.

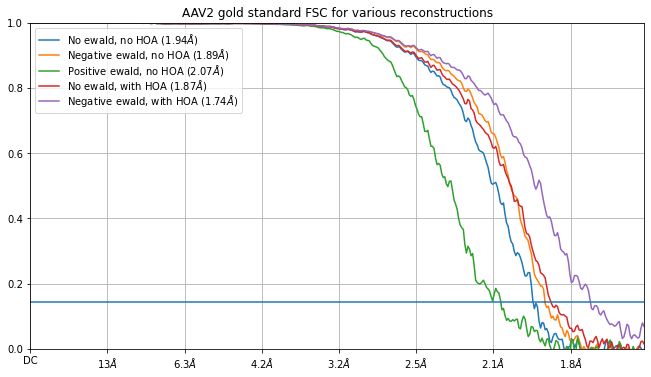
- A technique for Ewald Sphere Correction implemented in homogeneous refinement and reconstruction jobs allows for higher resolution reconstructions of larger proteins. Read a tutorial here. The new technique corrects for the Ewald Sphere during pose alignment and during back-projection, and also correctly estimates/refines higher-order CTF aberrations (HOA) while accounting for the Ewald Sphere.
- Anisotropic magnification can now be estimated during CTF refinement, and is corrected for in Refinement, Reconstruction, 3D Classification, and 3D Variability jobs.
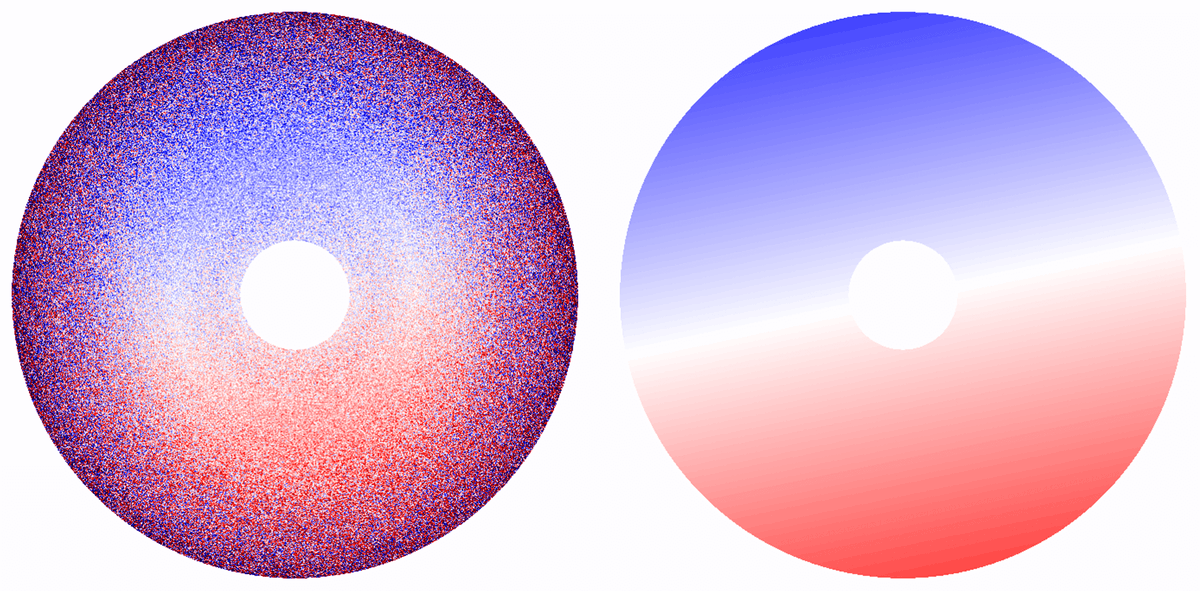
- Substantial speed and memory improvements to FSC calculations in all job types, speeding up FSC calculation by over 10x, and providing an overall speedup of up to 20% for homogeneous refinements. A benchmark refinement of 100,000 Ribosome particles at 360 pixel box size (EMPIAR-10028) takes only 5.2 minutes on a single NVIDIA V100 GPU.
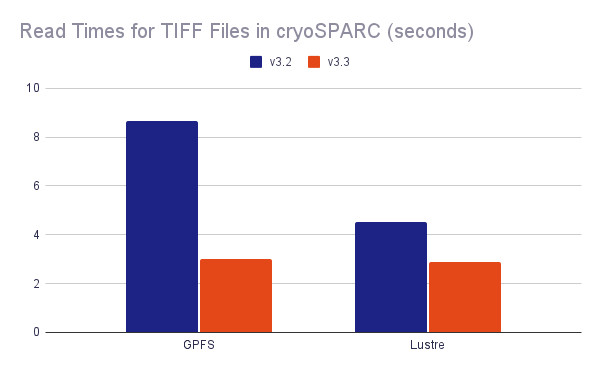
- Substantially improved TIFF read performance for some filesystems. Learn more.
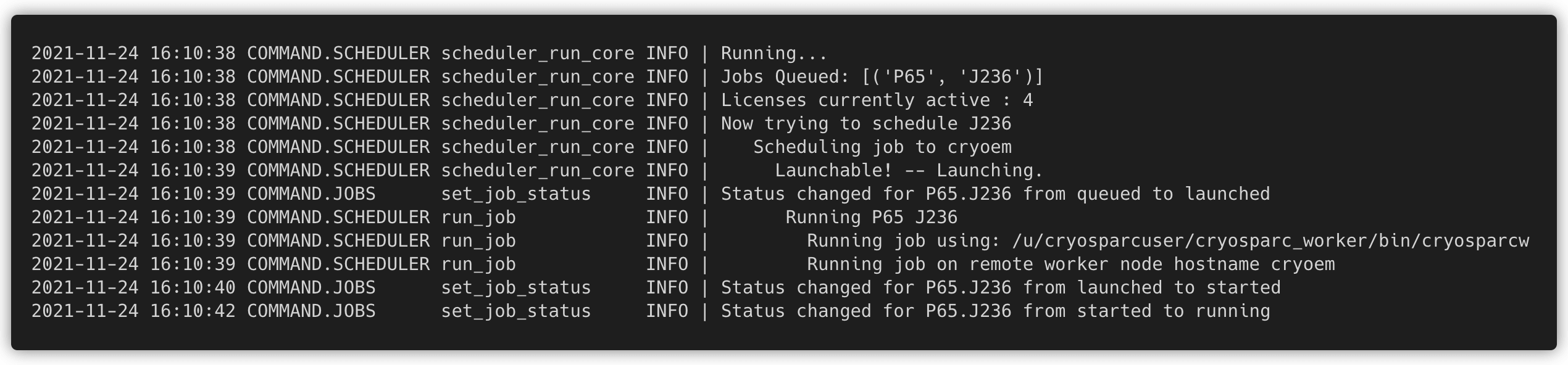
cryosparcm logprovides an improved log format and new filter flags.
- Import 3D Volumes now supports importing multiple volumes, including importing entries directly from EMDB.
- FSC Plots are now accompanied by an EMDB-friendly XML file.
- Improved performance in large projects. Faster Tree View updates and instantaneous input drag-and-drop connections.
- New Volume Alignment Tools utility, supporting re-centering and symmetry alignment of 3D volumes.
What’s Updated
- 3DVA Display job can now output particle sets corresponding to intermediate states.
- Added option to select a subset of variability components in 3DVA Display job.
- Volume Tools job now accepts a custom output filename. The default output name has also been updated to be more informative.
- Display path of job working directory at the top of the event log.
- Automatically delete SSD cache files that have not been used in more than 30 days during job caching steps.
- Specify path to custom Certificate Authority bundle for external SSL requests.
- CLI endpoint to safely update a project's directory (
update_project_directory(project_uid, new_project_dir)). - Ability to use common path field (vs. UID) to intersect or split in Exposure Sets Tool and Particle Sets Tool.
- CryoSPARC Live Data Management now supports executing a script upon a datatype's state change. Add
export CRYOSPARC_LIVE_DATA_MANAGEMENT_SCRIPT_ENABLE=trueandexport CRYOSPARC_LIVE_DATA_MANAGEMENT_SCRIPT_PATH=/path/to/script.shin cryosparc_master/config.sh to enable this feature. - Particle subtraction now respects refined higher-order aberrations.
- Added FSC mask auto-tightening to the standalone Validation (FSC) job.
- Extraction Box Size in Local Motion Correction now uses the input particle box size by default.
What’s Fixed
- Fixed contrast issues in Inspect Picks, Manual Picker and Curate Exposures jobs when viewing a micrograph that wasn't motion-corrected in cryoSPARC.
- Fixed issue in Volume Tools that caused ringing artefacts when downsampling masks.
- Added resolution parameter to low-pass filter templates in Blob and Template Picker jobs.
- Fixed
libtifferror when running the worker on a read-only file system. - Corrected volume flip in cryoSPARC Live 3D volume web viewer.
- Fixed a bug that caused stale notifications to persist after restarting cryoSPARC.
- Fixed a bug where it was not possible to queue a job into a new workspace.
- Fixed a bug where topics from the cryoSPARC Discussion Forum would not display on the dashboard.
- Updated the job queuing interface to clarify which nodes certain jobs are launched on. Interactive jobs are always launched on the master node. Import jobs are launched on the master node unless
CRYOSPARC_DISABLE_IMPORT_ON_MASTERis set to true, in which case import jobs can run on any lane/node selected. All other job types launch on a lane/node the user selects. - Fixed bug where symmetry alignment during refinement often aligned volumes incorrectly if they were already aligned.
- Fixed bug where volumes are improperly aligned to symmetry axes in helical refinement.
Learn how to update your cryoSPARC instance here
Have any questions about cryoSPARC or other applications for molecular dynamics?
Contact Exxact Today

CryoSPARC v3.3 Now Available
CryoSPARC Overview
CryoSPARC (Cryo-EM Single Particle Ab-initio Reconstruction and Classification) is a software package for processing cryo-electron microscopy (cryo-EM) single particle data, used in research and drug discovery.
As a complete solution for cryo-EM processing, cryoSPARC allows:
- Ultra-fast end-to-end processing of raw cryo-EM data and reconstruction of electron density maps, ready for ingestion into model building software
- Optimized algorithms and GPU acceleration at all stages, from pre-processing through particle picking, 2D particle classification, 3D ab-initio structure determination, high resolution refinement, and heterogeneity analysis
- Specialized and unique tools for therapeutically relevant targets, membrane proteins, continuously flexible structure
- Interactive, visual and iterative experimentation for even the most complex workflows
Interested in getting faster results?
Learn more about cryoSPARC Optimized GPU Workstations
CryoSPARC v3.3 Release
The newest update is packed with new features, performance optimizations and various stability enhancements.
Download the latest version here.
We highly recommend making a backup of your cryoSPARC database before updating.
Update instructions are available here.
What's New
- New 3D Classification BETA job can be used to quickly classify particles into many 3D classes, without re-alignment of particle poses and shifts. Sort out heterogeneity from million+ particle datasets with 100+ classes. Read a tutorial here.

- New ruler and scale bar allow for interactively determining the size of a particle in cryoSPARC Live.

- New Blob Picker Tuner determines optimal blob picker parameters from a minimal set of user-selected manual picks. Tunes blob size, shape, and picker score thresholds automatically. Read a tutorial here.

- A technique for Ewald Sphere Correction implemented in homogeneous refinement and reconstruction jobs allows for higher resolution reconstructions of larger proteins. Read a tutorial here. The new technique corrects for the Ewald Sphere during pose alignment and during back-projection, and also correctly estimates/refines higher-order CTF aberrations (HOA) while accounting for the Ewald Sphere.

- Anisotropic magnification can now be estimated during CTF refinement, and is corrected for in Refinement, Reconstruction, 3D Classification, and 3D Variability jobs.
- Substantial speed and memory improvements to FSC calculations in all job types, speeding up FSC calculation by over 10x, and providing an overall speedup of up to 20% for homogeneous refinements. A benchmark refinement of 100,000 Ribosome particles at 360 pixel box size (EMPIAR-10028) takes only 5.2 minutes on a single NVIDIA V100 GPU.
- Substantially improved TIFF read performance for some filesystems. Learn more.

cryosparcm logprovides an improved log format and new filter flags.

- Import 3D Volumes now supports importing multiple volumes, including importing entries directly from EMDB.
- FSC Plots are now accompanied by an EMDB-friendly XML file.
- Improved performance in large projects. Faster Tree View updates and instantaneous input drag-and-drop connections.
- New Volume Alignment Tools utility, supporting re-centering and symmetry alignment of 3D volumes.
What’s Updated
- 3DVA Display job can now output particle sets corresponding to intermediate states.
- Added option to select a subset of variability components in 3DVA Display job.
- Volume Tools job now accepts a custom output filename. The default output name has also been updated to be more informative.
- Display path of job working directory at the top of the event log.
- Automatically delete SSD cache files that have not been used in more than 30 days during job caching steps.
- Specify path to custom Certificate Authority bundle for external SSL requests.
- CLI endpoint to safely update a project's directory (
update_project_directory(project_uid, new_project_dir)). - Ability to use common path field (vs. UID) to intersect or split in Exposure Sets Tool and Particle Sets Tool.
- CryoSPARC Live Data Management now supports executing a script upon a datatype's state change. Add
export CRYOSPARC_LIVE_DATA_MANAGEMENT_SCRIPT_ENABLE=trueandexport CRYOSPARC_LIVE_DATA_MANAGEMENT_SCRIPT_PATH=/path/to/script.shin cryosparc_master/config.sh to enable this feature. - Particle subtraction now respects refined higher-order aberrations.
- Added FSC mask auto-tightening to the standalone Validation (FSC) job.
- Extraction Box Size in Local Motion Correction now uses the input particle box size by default.
What’s Fixed
- Fixed contrast issues in Inspect Picks, Manual Picker and Curate Exposures jobs when viewing a micrograph that wasn't motion-corrected in cryoSPARC.
- Fixed issue in Volume Tools that caused ringing artefacts when downsampling masks.
- Added resolution parameter to low-pass filter templates in Blob and Template Picker jobs.
- Fixed
libtifferror when running the worker on a read-only file system. - Corrected volume flip in cryoSPARC Live 3D volume web viewer.
- Fixed a bug that caused stale notifications to persist after restarting cryoSPARC.
- Fixed a bug where it was not possible to queue a job into a new workspace.
- Fixed a bug where topics from the cryoSPARC Discussion Forum would not display on the dashboard.
- Updated the job queuing interface to clarify which nodes certain jobs are launched on. Interactive jobs are always launched on the master node. Import jobs are launched on the master node unless
CRYOSPARC_DISABLE_IMPORT_ON_MASTERis set to true, in which case import jobs can run on any lane/node selected. All other job types launch on a lane/node the user selects. - Fixed bug where symmetry alignment during refinement often aligned volumes incorrectly if they were already aligned.
- Fixed bug where volumes are improperly aligned to symmetry axes in helical refinement.
Learn how to update your cryoSPARC instance here
Have any questions about cryoSPARC or other applications for molecular dynamics?
Contact Exxact Today




.jpg?format=webp)