
CryoSPARC Overview
CryoSPARC (Cryo-EM Single Particle Ab-initio Reconstruction and Classification) is a software package for processing cryo-electron microscopy (cryo-EM) single-particle data, used in research and drug discovery.
As a complete solution for cryo-EM processing, cryoSPARC allows:
- Ultra-fast end-to-end processing of raw cryo-EM data and reconstruction of electron density maps, ready for ingestion into model-building software
- Optimized algorithms and GPU acceleration at all stages, from pre-processing through particle picking, 2D particle classification, 3D ab-initio structure determination, high-resolution refinement, and heterogeneity analysis
- Specialized and unique tools for therapeutically relevant targets, membrane proteins, and continuously flexible structure
- Interactive, visual, and iterative experimentation for even the most complex workflows
Interested in getting faster results?
Learn more about cryoSPARC Optimized GPU Workstations
CryoSPARC v4.3 Release
The newest update is packed with new features, performance optimizations, and various stability enhancements.
New Features
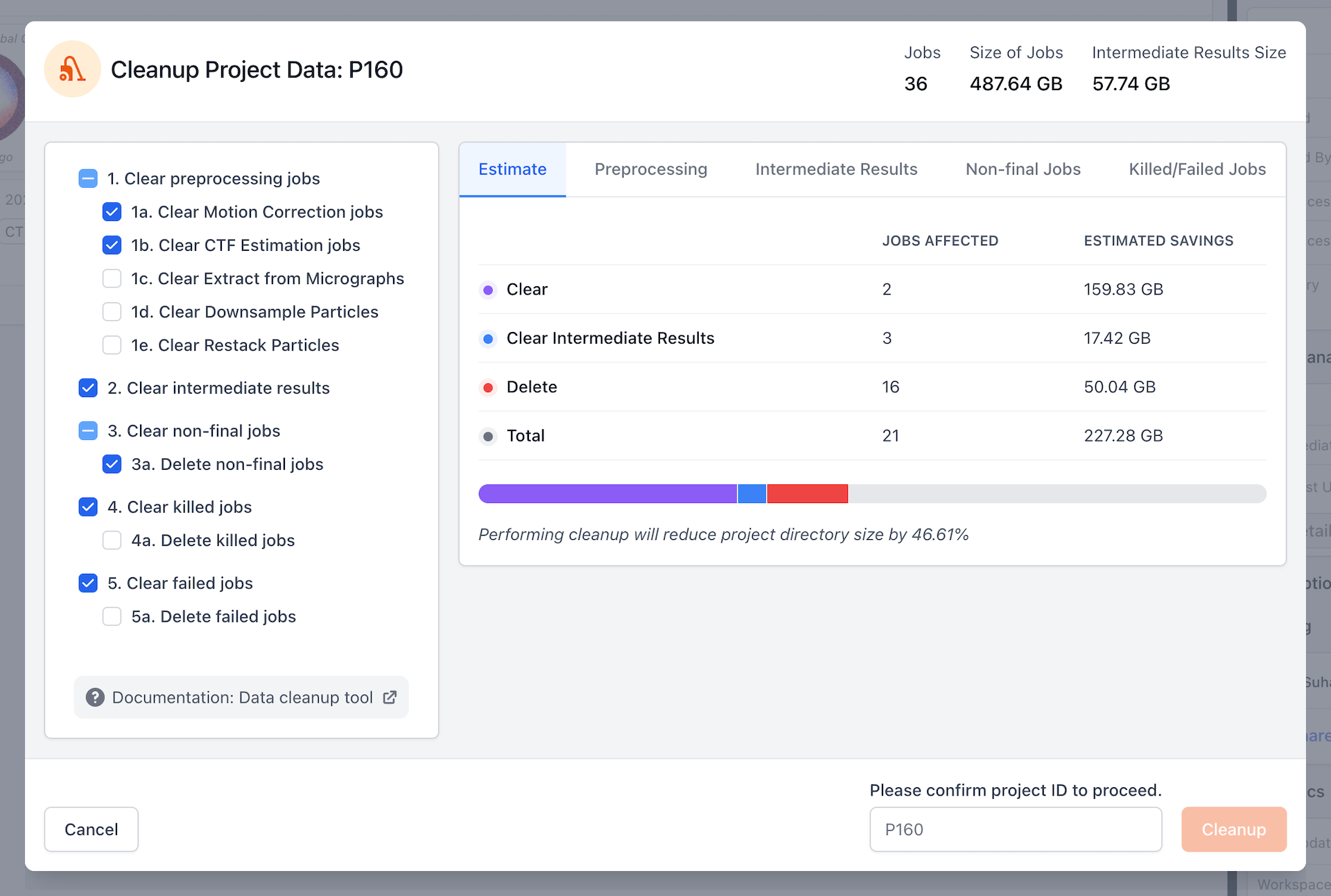
Data Cleanup Tools
- A new suite of tools to simplify the process of tracking and reducing disk space usage in CryoSPARC.
- Mark certain jobs as final and automatically clear everything except for jobs needed to replicate those results
- Compact CryoSPARC Live sessions, removing preprocessing data but storing particle locations and parameters needed to restore the session later
- Restack particles to remove filtered-out junk particles and then automatically clear deterministic preprocessing jobs that can be re-run later if needed
- Clear intermediate results of iterative jobs across a project or workspace
- Clear killed and failed jobs that take up disk space but can’t be further used
- Featuring a comprehensive Cleanup Data user interface, on-demand refreshing of project and job size statistics, data usage summaries in the project details sidebar, and new right-click menu options for selecting ancestors and descendants of jobs manually. See the CryoSPARC Data Cleanup guide for more details.
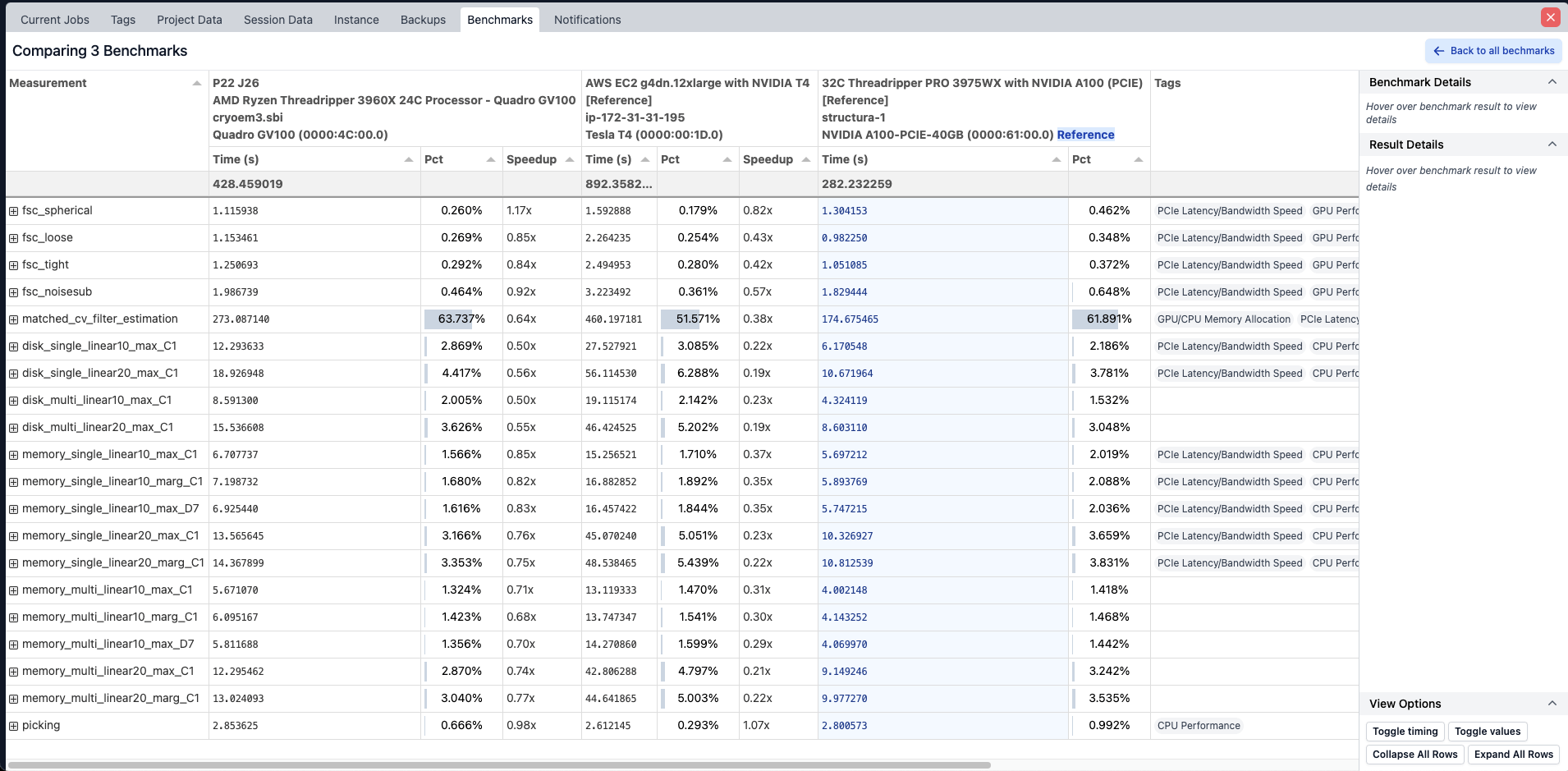
Performance Benchmarking Utilities
- A new system for measuring and comparing key aspects of CryoSPARC performance across CPU, File system, and GPU operations. The benchmarking jobs break down critical elements of CryoSPARC high performance code paths and measure detailed timings to shed light on which parts of a system may be bottlenecking performance.
- A new performance benchmark user interface allowing for visual comparison of different benchmark runs as well as reference benchmarks published along with CryoSPARC. The interface can display detailed timings from benchmarking jobs or job runtimes from full runs of the Extensive Validation (previously called Extensive Workflow) test set.
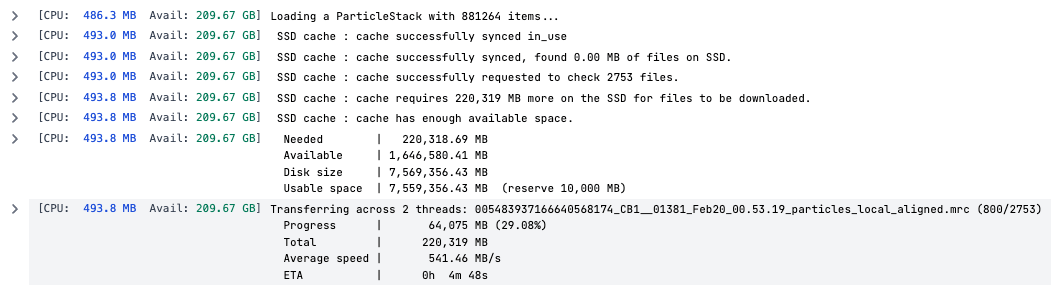
Multi-Threaded Particle Caching
Particle caching in CryoSPARC jobs now support multiple threads (default 2), increasing copy speed on certain filesystems. To modify, specify CRYOSPARC_CACHE_NUM_THREADS in cryosparc_worker/config.sh. Read the guide for more information.
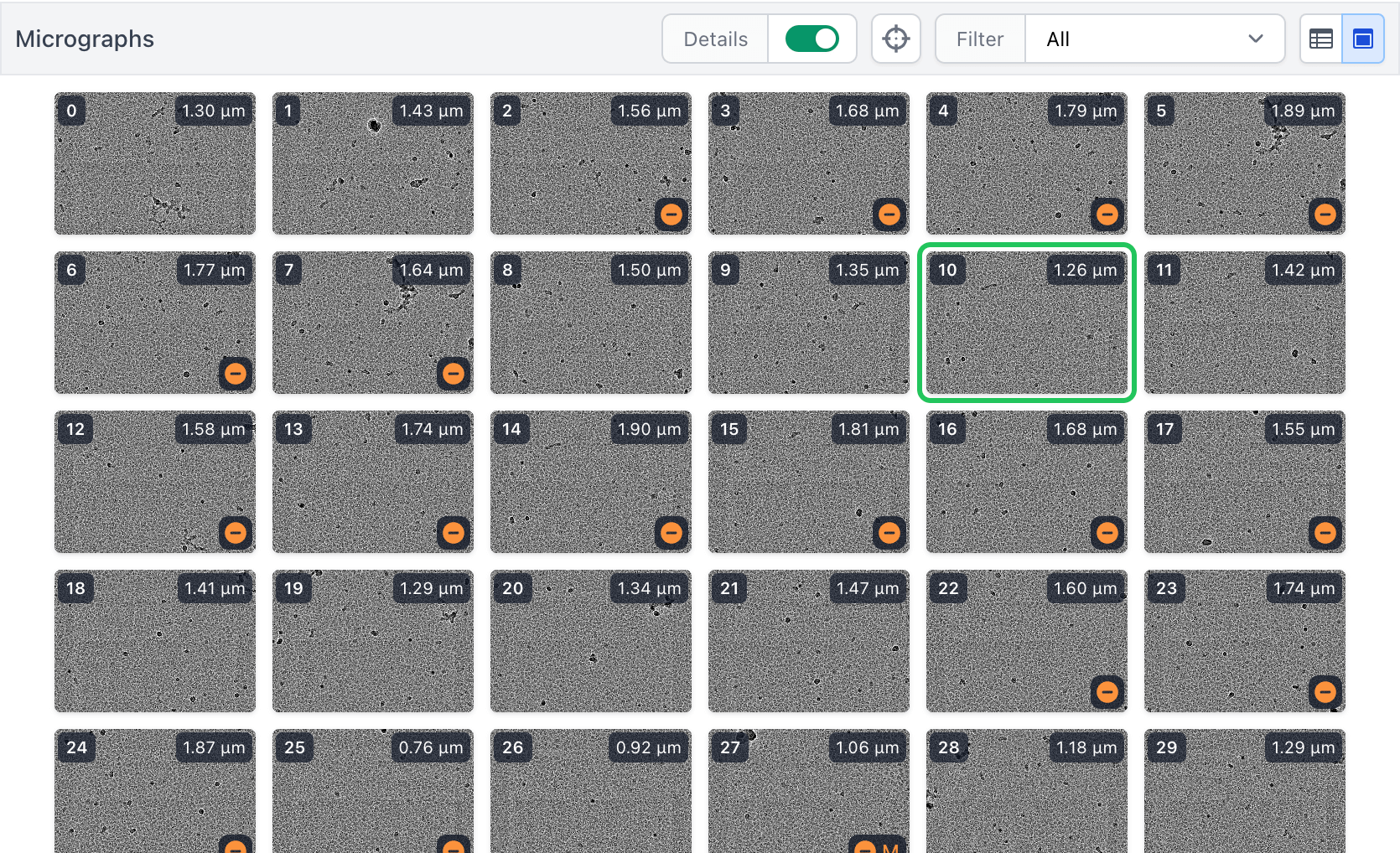
Interactive Job Improvements
- New plots, displays, and interactive tools across all interactive jobs, including a new exposure image grid view in the Exposure Curation job for ease of assessing and manually curating exposures, and the ability to set threshold parameters when starting the job to bypass interaction and automatically filter data.
- Several usability enhancements, including:
- Extraction is now optional in the manual picker
- Datasets without pick statistics can be used in Inspect Picks
- Charts in Exposure Curation remember their expand/collapse state when reloaded
- Exposure Curation now displays particle picks if available
- Optimized micrograph image loading for faster response times
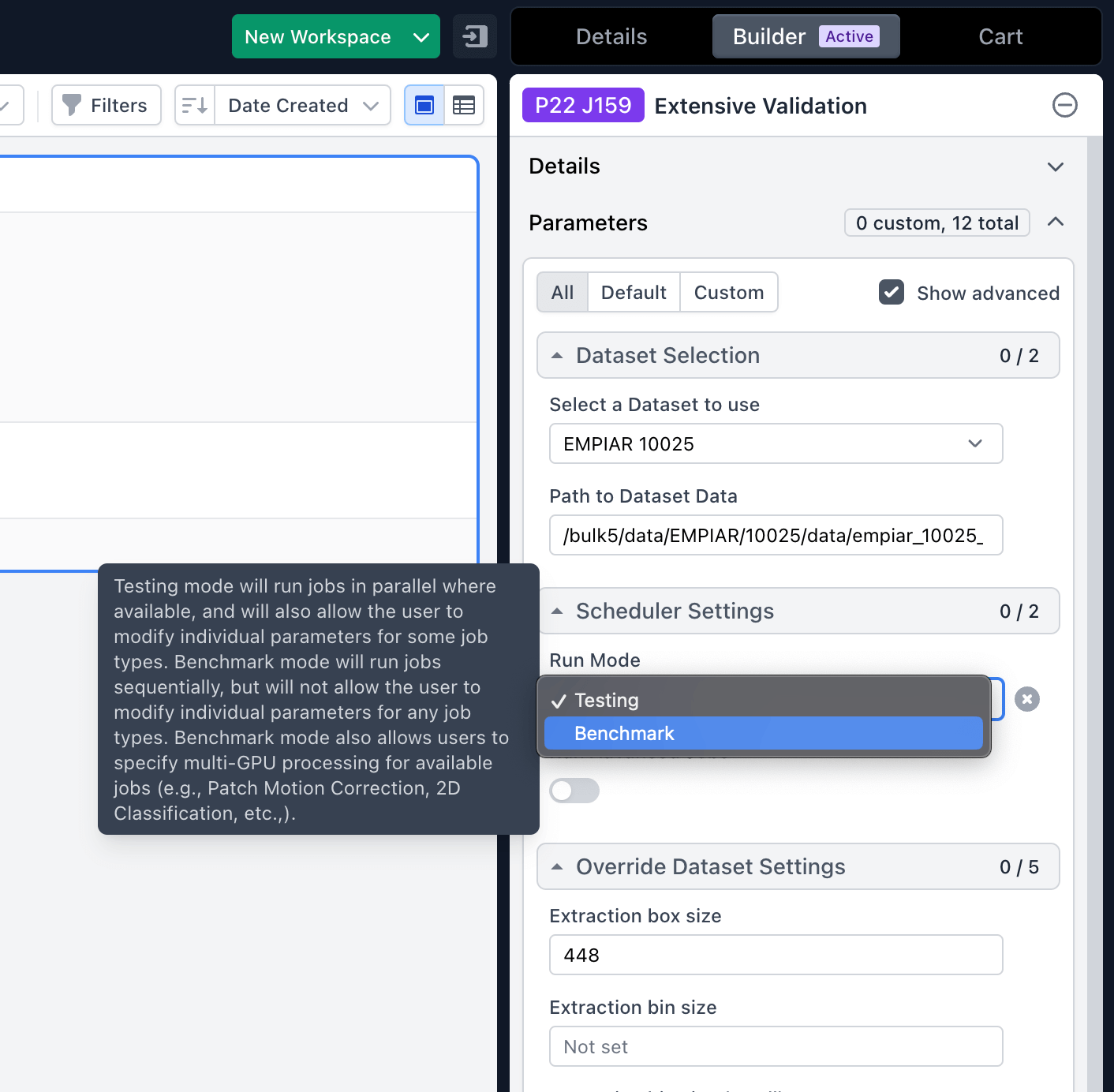
Extensive Validation
Extensive Validation (previously called Extensive Workflow) job extended to support multiple datasets and also has a new “Benchmark” mode where timing results are reported and browsable using the new Performance Benchmarking system.
The cryosparcm compact command can reduce the amount of disk space used by the MongoDB database. This, along with other steps, may be useful for reducing used storage space in instances with large database sizes. See the Guide: Reduce Database Size for more details.
Updates and Fixes
Updates
- Interactive job improvements:
- Flipped Select 2D resolution threshold selection option for clarity
- Added total picks across all exposures as info badge in Manual Picker top bar
- Added filter to Select 2D job to allow showing of only selected or unselected classes (sorting options can be applied to filtered classes)
- Added info badge to Inspect Picks job to display the total number of particles within the applied thresholds
- Manual Picker and Inspect Picks jobs now display all available data fields in the micrograph table
- Added an exposure image preview when hovering exposure table rows in interactive jobs
- Individual tab has been renamed to diagnostics in Exposure Curation job for clarity of function
- Thresholds of 0 now filter micrographs correctly in Exposure Curation job
- Support added for right clicking to remove picks in Manual Picker job
- Non-uniform Refinement implementation speedups up to 15% on certain GPUs and box sizes.
- Import Particles job is now 50-70% faster.
- Improved speed of the caching system by reducing the number of file lookups, and improved logging of the cache system while a job is running.
- CTFFIND4 is d to v4.1.14.
- Import and utility jobs that previously ran only on the master node can now optionally be launched on any worker node or cluster lane. The CRYOSPARC_DISABLE_IMPORT_ON_MASTER environment variable is no longer used.
- Added star button to top navigation bar in browse system to allow starring a workspace from the jobs page.
- Quick access panel will now remember selections on a user basis rather than a browser tab basis.
- The load all jobs view option is now saved per-user rather than based on the browser.
- Event log view options have been moved into a new view options menu to minimize interface clutter.
- Sessions can now be marked as completed from the session browser view.
- Archived and detached projects no longer show within the session browser.
- An Additional spotlight action added to navigate to the job history page.
- The ‘view job’ button in the sidebar can be CTRL / CMD clicked to open a new tab
- For job types that take random seed parameters, the random seed that was used is now saved in the input parameters of the job for easy reuse. The random seed will stay consistent if the job is cleared and re-run.
- 2D Classification, 3D Classification, and 3D Variability Analysis jobs will now produce intermediate results at each iteration, but will delete intermediate results from previous iterations as each new iteration completes. This behaviour can be turned off with a project-level or job-level parameter that will keep all intermediate results.
- Jobs that fail to report heartbeat to the master node within the required timeout but continue running in the background will now be killed on their worker node, rather than being allowed to continue running after having been marked as failed due to missing heartbeat.
- Worker installation job launch test (cryosparcm test workers) timeout increased to 2 minutes from 10 seconds.
- The config.sh file with environment variables will now be included in the cryosparcm snaplogs bundle of logs.
- CryoSPARC running version is now available in the Job PDF download.
- MongoDB database journaling can be enabled by adding export CRYOSPARC_MONGO_EXTRA_FLAGS="" to cryosparc_master/config.sh.
Fixes
- Topaz Extract job no longer extracts particles from denoised micrographs by default.
- 3DFlex Generate job no longer ignores the Number of Frames parameter.
- Fixed unnecessary AssertionError in Flex Mesh Prep job when pixel size between input mask and volume match but have rounding error difference
- Heterogenous Refinement job no longer fails with AttributeError when “Refinement box size (Voxels)” parameter passed None as a value.
- Worker installation test (cryosparcm test workers) no longer fails when worker target does not have SSD configured.
- CryoSPARC Live Session compute configuration parameters no longer fails to be changed if a worker lane that was previously selected is removed from the CryoSPARC instance.
- Fixed select confirmation appearance in Edge browser on Windows.
- Fixed bug where Job PDF would show incorrect parameter values in some cases.
- Fixed issue where PDF event log download was crashing due to control characters being interpreted incorrectly during text cleaning.
- Corrected sorting order of the job event log PDF.
- Added optimistic job count to stop the user interface loading state from glitching when deleting a workspace job when few exist.
- Resolved error where Import Result Group fails to import Topaz jobs correctly, causing downstream jobs to fail.
- Resolved error where Import 3D Volumes job fails with “Unable to connect to EMDB server” when trying to load volumes from EMBD.
- Allow retrieving job and workspace documents from the CLI without errors.
- Improved handling of missing job parent in tree view.
- Live no longer stops finding new exposures until the next restart.
Learn how to update your cryoSPARC instance here
Drive you cryo-EM workloads with a purpose built CryoSPARC workstation, server, or cluster. Exxact is highly experienced in delivering the best HPC solution for scientific computing. Contact us today to learn more today.

CryoSPARC v4.3 Release Notes
CryoSPARC Overview
CryoSPARC (Cryo-EM Single Particle Ab-initio Reconstruction and Classification) is a software package for processing cryo-electron microscopy (cryo-EM) single-particle data, used in research and drug discovery.
As a complete solution for cryo-EM processing, cryoSPARC allows:
- Ultra-fast end-to-end processing of raw cryo-EM data and reconstruction of electron density maps, ready for ingestion into model-building software
- Optimized algorithms and GPU acceleration at all stages, from pre-processing through particle picking, 2D particle classification, 3D ab-initio structure determination, high-resolution refinement, and heterogeneity analysis
- Specialized and unique tools for therapeutically relevant targets, membrane proteins, and continuously flexible structure
- Interactive, visual, and iterative experimentation for even the most complex workflows
Interested in getting faster results?
Learn more about cryoSPARC Optimized GPU Workstations
CryoSPARC v4.3 Release
The newest update is packed with new features, performance optimizations, and various stability enhancements.
New Features
Data Cleanup Tools
- A new suite of tools to simplify the process of tracking and reducing disk space usage in CryoSPARC.
- Mark certain jobs as final and automatically clear everything except for jobs needed to replicate those results
- Compact CryoSPARC Live sessions, removing preprocessing data but storing particle locations and parameters needed to restore the session later
- Restack particles to remove filtered-out junk particles and then automatically clear deterministic preprocessing jobs that can be re-run later if needed
- Clear intermediate results of iterative jobs across a project or workspace
- Clear killed and failed jobs that take up disk space but can’t be further used
- Featuring a comprehensive Cleanup Data user interface, on-demand refreshing of project and job size statistics, data usage summaries in the project details sidebar, and new right-click menu options for selecting ancestors and descendants of jobs manually. See the CryoSPARC Data Cleanup guide for more details.

Performance Benchmarking Utilities
- A new system for measuring and comparing key aspects of CryoSPARC performance across CPU, File system, and GPU operations. The benchmarking jobs break down critical elements of CryoSPARC high performance code paths and measure detailed timings to shed light on which parts of a system may be bottlenecking performance.
- A new performance benchmark user interface allowing for visual comparison of different benchmark runs as well as reference benchmarks published along with CryoSPARC. The interface can display detailed timings from benchmarking jobs or job runtimes from full runs of the Extensive Validation (previously called Extensive Workflow) test set.

Multi-Threaded Particle Caching
Particle caching in CryoSPARC jobs now support multiple threads (default 2), increasing copy speed on certain filesystems. To modify, specify CRYOSPARC_CACHE_NUM_THREADS in cryosparc_worker/config.sh. Read the guide for more information.

Interactive Job Improvements
- New plots, displays, and interactive tools across all interactive jobs, including a new exposure image grid view in the Exposure Curation job for ease of assessing and manually curating exposures, and the ability to set threshold parameters when starting the job to bypass interaction and automatically filter data.
- Several usability enhancements, including:
- Extraction is now optional in the manual picker
- Datasets without pick statistics can be used in Inspect Picks
- Charts in Exposure Curation remember their expand/collapse state when reloaded
- Exposure Curation now displays particle picks if available
- Optimized micrograph image loading for faster response times

Extensive Validation
Extensive Validation (previously called Extensive Workflow) job extended to support multiple datasets and also has a new “Benchmark” mode where timing results are reported and browsable using the new Performance Benchmarking system.

The cryosparcm compact command can reduce the amount of disk space used by the MongoDB database. This, along with other steps, may be useful for reducing used storage space in instances with large database sizes. See the Guide: Reduce Database Size for more details.
Updates and Fixes
Updates
- Interactive job improvements:
- Flipped Select 2D resolution threshold selection option for clarity
- Added total picks across all exposures as info badge in Manual Picker top bar
- Added filter to Select 2D job to allow showing of only selected or unselected classes (sorting options can be applied to filtered classes)
- Added info badge to Inspect Picks job to display the total number of particles within the applied thresholds
- Manual Picker and Inspect Picks jobs now display all available data fields in the micrograph table
- Added an exposure image preview when hovering exposure table rows in interactive jobs
- Individual tab has been renamed to diagnostics in Exposure Curation job for clarity of function
- Thresholds of 0 now filter micrographs correctly in Exposure Curation job
- Support added for right clicking to remove picks in Manual Picker job
- Non-uniform Refinement implementation speedups up to 15% on certain GPUs and box sizes.
- Import Particles job is now 50-70% faster.
- Improved speed of the caching system by reducing the number of file lookups, and improved logging of the cache system while a job is running.
- CTFFIND4 is d to v4.1.14.
- Import and utility jobs that previously ran only on the master node can now optionally be launched on any worker node or cluster lane. The CRYOSPARC_DISABLE_IMPORT_ON_MASTER environment variable is no longer used.
- Added star button to top navigation bar in browse system to allow starring a workspace from the jobs page.
- Quick access panel will now remember selections on a user basis rather than a browser tab basis.
- The load all jobs view option is now saved per-user rather than based on the browser.
- Event log view options have been moved into a new view options menu to minimize interface clutter.
- Sessions can now be marked as completed from the session browser view.
- Archived and detached projects no longer show within the session browser.
- An Additional spotlight action added to navigate to the job history page.
- The ‘view job’ button in the sidebar can be CTRL / CMD clicked to open a new tab
- For job types that take random seed parameters, the random seed that was used is now saved in the input parameters of the job for easy reuse. The random seed will stay consistent if the job is cleared and re-run.
- 2D Classification, 3D Classification, and 3D Variability Analysis jobs will now produce intermediate results at each iteration, but will delete intermediate results from previous iterations as each new iteration completes. This behaviour can be turned off with a project-level or job-level parameter that will keep all intermediate results.
- Jobs that fail to report heartbeat to the master node within the required timeout but continue running in the background will now be killed on their worker node, rather than being allowed to continue running after having been marked as failed due to missing heartbeat.
- Worker installation job launch test (cryosparcm test workers) timeout increased to 2 minutes from 10 seconds.
- The config.sh file with environment variables will now be included in the cryosparcm snaplogs bundle of logs.
- CryoSPARC running version is now available in the Job PDF download.
- MongoDB database journaling can be enabled by adding export CRYOSPARC_MONGO_EXTRA_FLAGS="" to cryosparc_master/config.sh.
Fixes
- Topaz Extract job no longer extracts particles from denoised micrographs by default.
- 3DFlex Generate job no longer ignores the Number of Frames parameter.
- Fixed unnecessary AssertionError in Flex Mesh Prep job when pixel size between input mask and volume match but have rounding error difference
- Heterogenous Refinement job no longer fails with AttributeError when “Refinement box size (Voxels)” parameter passed None as a value.
- Worker installation test (cryosparcm test workers) no longer fails when worker target does not have SSD configured.
- CryoSPARC Live Session compute configuration parameters no longer fails to be changed if a worker lane that was previously selected is removed from the CryoSPARC instance.
- Fixed select confirmation appearance in Edge browser on Windows.
- Fixed bug where Job PDF would show incorrect parameter values in some cases.
- Fixed issue where PDF event log download was crashing due to control characters being interpreted incorrectly during text cleaning.
- Corrected sorting order of the job event log PDF.
- Added optimistic job count to stop the user interface loading state from glitching when deleting a workspace job when few exist.
- Resolved error where Import Result Group fails to import Topaz jobs correctly, causing downstream jobs to fail.
- Resolved error where Import 3D Volumes job fails with “Unable to connect to EMDB server” when trying to load volumes from EMBD.
- Allow retrieving job and workspace documents from the CLI without errors.
- Improved handling of missing job parent in tree view.
- Live no longer stops finding new exposures until the next restart.
Learn how to update your cryoSPARC instance here
Drive you cryo-EM workloads with a purpose built CryoSPARC workstation, server, or cluster. Exxact is highly experienced in delivering the best HPC solution for scientific computing. Contact us today to learn more today.




.jpg?format=webp)